CLIF Data Dictionary
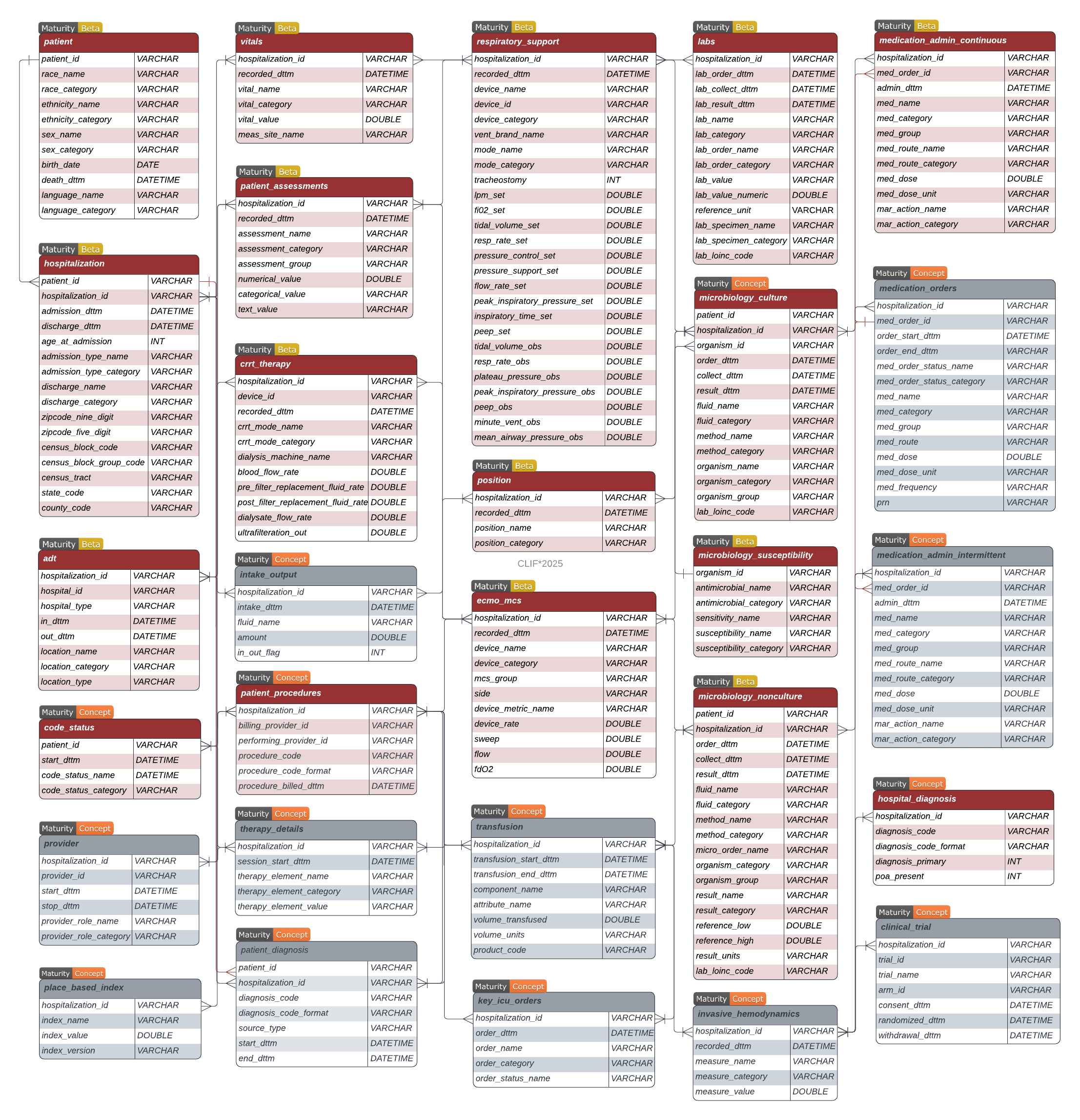
The CLIF Data Dictionary serves as a comprehensive guide to the Common Longitudinal ICU data Format, detailing the structure and purpose of each table within the framework. Designed to standardize and harmonize electronic health record data across multiple institutions, the dictionary outlines the entity-relationship model, variable definitions, and permissible values.
Beta Tables
The table purpose, structure, and field names for beta tables is complete and used in at least one federated CLIF project. The minimum Common ICU Data Elements (mCIDE) for category variables is defined. Actively testing the table’s practical use in projects. Breaking changes unlikely, but backward compatible updates in future minor versions possible
adt Beta
The admission, discharge, and transfer (adt) table is a start-stop longitudinal dataset that contains information about each patient’s movement within the hospital. It also has a hospital_id field to distinguish between different hospitals within a health system.
| Column | Type | Description | Permissible Values |
|---|---|---|---|
| hospitalization_id | VARCHAR | ID variable for each patient encounter | No restriction |
| hospital_id | VARCHAR | Assign a unique ID to each hospital within a healthsystem | No restriction |
| hospital_type | VARCHAR | Maps hospital_id to a standardized list of hospital types | academic, community, LTACH |
| in_dttm | DATETIME | Start date and time at a particular location. All datetime variables must be timezone-aware and set to UTC. | Datetime format should be YYYY-MM-DD HH:MM:SS+00:00 (UTC) |
| out_dttm | DATETIME | End date and time at a particular location. All datetime variables must be timezone-aware and set to UTC. | Datetime format should be YYYY-MM-DD HH:MM:SS+00:00 (UTC) |
| location_name | VARCHAR | Location of the patient inside the hospital. This field is used to store the patient location from the source data. It is not used for analysis. | No restriction |
| location_category | VARCHAR | Maps location_name to a standardized list of ADT location categories | ed, ward, stepdown, icu, procedural, l&d, hospice, psych, rehab, radiology, dialysis, other |
| location_type | VARCHAR | Maps ICU type to a standardized list of ICU categories | List of ICU categories in CLIF |
Notes:
- ADT represents the patient’s physical location, NOT the patient “status”.
- Procedural areas and operating rooms should be mapped to
Procedural. Pre/Intra/Post-procedural/OR EHR data (such as anesthesia flowsheet records from Labs, Vitals, Scores, Respiratory Support) are not currently represented in CLIF.
Example:
| hospitalization_id | hospital_id | hospital_type | in_dttm | out_dttm | location_name | location_category | location_type |
|---|---|---|---|---|---|---|---|
| 20010012 | ABC | academic | 2024-12-01 10:00:00+00:00 UTC | 2024-12-01 14:00:00+00:00 UTC | B06F | icu | general_icu |
| 20010012 | ABC | academic | 2024-12-01 14:30:00+00:00 UTC | 2024-12-02 08:00:00+00:00 UTC | B78D | ward | |
| 20010015 | ABC | academic | 2024-11-30 16:45:00+00:00 UTC | 2024-12-01 12:00:00+00:00 UTC | B06T | icu | general_icu |
| 20010015 | ABC | academic | 2024-12-01 12:30:00+00:00 UTC | 2024-12-02 07:00:00+00:00 UTC | N23E | procedural | |
| 20010020 | EFG | community | 2024-11-28 09:00:00+00:00 UTC | 2024-11-29 17:00:00+00:00 UTC | B78D | ward |
code status Beta
This table contains only code status orders placed by clinicians. It is NOT equivalent to the code status display name in the EMR.
| Column | Type | Description | Permissible Values |
|---|---|---|---|
| patient_id | VARCHAR | Unique identifier for each patient, presumed to be a distinct individual. | No restriction |
| start_dttm | DATETIME | The date and time when the specific code status was initiated | Datetime format should be YYYY-MM-DD HH:MM:SS+00:00 (UTC) |
| code_status_name | VARCHAR | The name/description of the code status | No restriction |
| code_status_category | VARCHAR | Categorical variable specifying the code status during the hospitalization | DNR, DNAR, UDNR, DNR/DNI, DNAR/DNI, AND, Full, Presume Full, Other |
Notes:
- The
code_status_categoryset of permissible values is under development
Example:
| patient_id | start_dttm | code_status_name | code_status_category |
|---|---|---|---|
| 123451 | 2024-12-01 08:30:00+00:00 UTC | Do Not Resuscitate | DNR |
| 123452 | 2024-12-02 14:00:00+00:00 UTC | Do Not Intubate | DNR/DNI |
| 123451 | 2024-12-03 10:15:00+00:00 UTC | Full Code | Full |
crrt therapy Beta
The crrt_therapy table captures Continuous Renal Replacement Therapy (CRRT) data, including different CRRT modalities, operational parameters, and fluid exchange details. The intermittent HD, peritoneal dialysis, PERT, and SLED tables are under development.
| Column | Type | Description | Permissible Values |
|---|---|---|---|
| hospitalization_id | VARCHAR | ID variable for each patient encounter | No restriction |
| device_id | VARCHAR | Unique ID of the individual dialysis machine used (e.g., machine ACZ3RV91). Distinct from dialysis_machine_name, which stores the brand/model. | No restriction |
| recorded_dttm | DATETIME | Timestamp when CRRT parameters were recorded | Datetime format should be YYYY-MM-DD HH:MM:SS+00:00 (UTC) |
| crrt_mode_name | VARCHAR | Name of CRRT mode (e.g., CVVHDF) | No restriction |
| crrt_mode_category | VARCHAR | Standardized CRRT mode categories | scuf, cvvh, cvvhd, cvvhdf, avvh |
| dialysis_machine_name | VARCHAR | Brand name for the dialysis machine | No restriction |
| blood_flow_rate | FLOAT | Rate of blood flow through the CRRT circuit (mL/min) | 150-350 |
| pre_filter_replacement_fluid_rate | FLOAT | Rate of pre-filter replacement fluid infusion (mL/hr) | 0-10000 |
| post_filter_replacement_fluid_rate | FLOAT | Rate of post-filter replacement fluid infusion (mL/hr) | 0-10000 |
| dialysate_flow_rate | FLOAT | Flow rate of dialysate solution (mL/hr) | 0-10000 |
| ultrafiltration_out | FLOAT | Net ultrafiltration output (mL/hr) | 0-500 |
Notes:
- SCUF: Slow Continuous Ultrafiltration
- CVVH: Continuous Veno-Venous Hemofiltration
- CVVHD: Continuous Veno-Venous Hemodialysis
- CVVHDF: Continuous Venous-Venous Hemodiafiltration
- AVVH: Accelerated Veno-venous Hemofiltration also called ARRT or PIIRT
CRRT Modalities and Parameter Usage:
| CRRT Modality | Blood Flow Rate | Pre-Filter Replacement Rate | Post-Filter Replacement Rate | Dialysate Flow Rate | Ultrafiltration Out |
|---|---|---|---|---|---|
| SCUF | Required | Not Used | Not Used | Not Used | Required |
| CVVH | Required | Required | Required | Not Used | Required |
| CVVHD | Required | Not Used | Not Used | Required | Required |
| CVVHDF | Required | Required | Required | Required | Required |
| AVVH (VVH) | Required | May Be Used | May Be Used | Not Used | Required |
| AVVH (VVHD) | Required | Not Used | Not Used | May Be Used | Required |
| AVVH (VVHF) | Required | May Be Used | May Be Used | May Be Used | Required |
Example:
| hospitalization_id | device_id | recorded_dttm | crrt_mode_name | crrt_mode_category | dialysis_machine_name | blood_flow_rate | pre_filter_replacement_fluid_rate | post_filter_replacement_fluid_rate | dialysate_flow_rate | ultrafiltration_out |
|---|---|---|---|---|---|---|---|---|---|---|
| 201 | J0 | 2024-02-15 07:00:00+00:00 UTC | CVVHDF | CVVHDF | NxStage by Baxter | 200.0 | 1000.0 | 500.0 | 800.0 | 1500.0 |
| 202 | J0 | 2024-02-16 09:15:00+00:00 UTC | CVVH | CVVH | NxStage by Baxter | 180.0 | 1200.0 | 300.0 | NA | 1300.0 |
| 203 | J0 | 2024-02-17 11:45:00+00:00 UTC | SCUF | SCUF | NxStage by Baxter | 150.0 | NA | NA | NA | 800.0 |
hospital diagnosis Beta
Finalized billing diagnosis codes for hospital reimbursement, e.g. calculation of a Diagnosis Related Group (DRG). These diagnoses also do not have timestamps, as they are often finalized after discharge. The hospital_diagnosis table is appropriate for calculation of comorbidity scores but should not be used as input features into a prediction model for an inpatient event.
All other diagnosis codes for a patient are included under concept table patient_diagnosis which has start and end timestamps.
| Column | Type | Description | Permissible Values |
|---|---|---|---|
| hospitalization_id | VARCHAR | ID variable for each patient encounter | Must match a hospitalization_id in the hospitalization table |
| diagnosis_code | VARCHAR | ICD diagnosis code | Valid ICD-9-CM or ICD-10-CM code |
| diagnosis_code_format | VARCHAR | Format of the code | ICD10CM or ICD9CM |
| diagnosis_primary | INT | Type of diagnosis: 1 = primary, 0 = secondary. If diagnoses are ranked, any rank of 2 or above is considered secondary. | 0, 1 |
| poa_present | INT | Indicator if the diagnosis was present on admission. Only two options are allowed: 1 = Yes (present on admission), 0 = No (not present on admission). No other values (such as Exempt, Unknown, or Unspecified) are permitted. | 0, 1 |
Example:
| hospitalization_id | diagnosis_code | diagnosis_code_format | diagnosis_primary | poa_present |
|---|---|---|---|---|
| 20010012 | I10 | ICD10CM | 1 | 1 |
| 20010012 | E11.9 | ICD10CM | 0 | 0 |
| 20010015 | 250.00 | ICD9CM | 1 | 1 |
| 20010015 | 401.9 | ICD9CM | 0 | 0 |
| 20010020 | J45.909 | ICD10CM | 1 | 1 |
| 20010020 | 530.81 | ICD9CM | 0 | 1 |
hospitalization Beta
The hospitalization table contains information about each hospitalization event. Each row in this table represents a unique hospitalization event for a patient. This table is inspired by the visit_occurance OMOP table but is specific to inpatient hospitalizations (including those that begin in the emergency room).
| Column | Type | Description | Permissible Values |
|---|---|---|---|
| patient_id | VARCHAR | Unique identifier for each patient, presumed to be a distinct individual. | No restriction |
| hospitalization_id | VARCHAR | ID variable for each patient encounter | No restriction |
| hospitalization_joined_id | VARCHAR | Unique identifier for each continuous inpatient stay in a health system which may span different hospitals (Optional) | No restriction |
| admission_dttm | DATETIME | Date and time the patient is admitted to the hospital. All datetime variables must be timezone-aware and set to UTC | Datetime format should be YYYY-MM-DD HH:MM:SS+00:00 (UTC) |
| discharge_dttm | DATETIME | Date and time the patient is discharged from the hospital. All datetime variables must be timezone-aware and set to UTC | Datetime format should be YYYY-MM-DD HH:MM:SS+00:00 (UTC) |
| age_at_admission | INT | Age of the patient at the time of admission, in years | No restriction |
| admission_type_name | VARCHAR | Type of inpatient admission. Original string from the source data | e.g. Direct admission, Transfer, Pre-op surgical |
| admission_type_category | VARCHAR | Admission disposition mapped to mCIDE categories | List of admission type categories in CLIF |
| discharge_name | VARCHAR | Original discharge disposition name string recorded in the raw data | No restriction, e.g. home |
| discharge_category | VARCHAR | Maps discharge_name to a standardized list of discharge categories | Home, Skilled Nursing Facility (SNF), Expired, Acute Inpatient Rehab Facility, Hospice, Long Term Care Hospital (LTACH), Acute Care Hospital, Group Home, Chemical Dependency, Against Medical Advice (AMA), Assisted Living, Still Admitted, Missing, Other, Psychiatric Hospital, Shelter, Jail |
| zipcode_nine_digit | VARCHAR | Patient 9 digit zip code, used to link with other indices such as ADI and SVI | No restriction |
| zipcode_five_digit | VARCHAR | Patient 5 digit zip code, used to link with other indices such as ADI and SVI | No restriction |
| census_block_code | VARCHAR | 15 digit FIPS code | No restriction |
| census_block_group_code | VARCHAR | 12 digit FIPS code | No restriction |
| census_tract | VARCHAR | Full 11 digit FIPS code. Eg. 13089022404 census_tract is the state (13) + the county (089) + the census tract (022404). | No restriction |
| state_code | VARCHAR | 2 digit FIPS code | No restriction |
| county_code | VARCHAR | Full 5 digit FIPS code. Eg. 13089 county_code is the state (13) + the county (089). | No restriction |
| fips_version | VARCHAR | Year of the Census geography definitions used for the FIPS codes (e.g., 2010, 2020), indicating the tract and boundary set in effect at that time | 2000,2010, 2020 |
Notes:
- If a patient is discharged to Home/Hospice, then
discharge_category == Hospice. - Mortality Outcomes: Mortality is identified by
discharge_category == "Expired". Some studies also includedischarge_category == "Hospice"(i.e., both “Expired” and “Hospice”) when defining death outcomes. - The geographical indicators (
zipcode_nine_digit,zipcode_five_digit,census_block_code,census_block_group_code,census_tract,state_code,county_code) should be added if they are available in your source dataset.zipcode_nine_digitis preferred overzipcode_five_digit, andcensus_block_codeis ideal for census based indicators. The choice of geographical indicators may differ depending on the project. - If a patient is transferred between different hospitals within a health system, a new
hospitalization_idshould be created. - If a patient is initially seen in an ER in hospital A and then admitted to inpatient status in hospital B, one
hospitalization_idshould be created for data from both stays. - A
hospitalization_joined_idcan also be created from a CLIF table from contiguoushospitalization_ids. - Geo-based Indices (ADI, SVI):
- ADI: Calculate the Area Deprivation Index at the census block-group level. Provide
census_block_group_codewhen possible, orzipcode_nine_digit(9-digit ZIP) that can be cross-walked to a block group. Avoid using 5-digit ZIP or census-tract values for ADI—they are not validated and will add error. - SVI: The Social Vulnerability Index is published at
census_tract(full 11-digit FIPS).
- ADI: Calculate the Area Deprivation Index at the census block-group level. Provide
Example:
| patient_id | hospitalization_id | hospitalization_joined_id | admission_dttm | discharge_dttm | age_at_admission | admission_type_name | admission_type_category | discharge_name | discharge_category | zipcode_five_digit | zipcode_nine_digit | census_block_group_code | latitude | longitude | fips_version |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 101001 | 12345678 | 22334455 | 2024-11-01 08:15:00+00:00 UTC | 2024-11-04 14:30:00+00:00 UTC | 65 | Direct admission | Inpatient | Discharged to Home or Self Care (Routine Discharge) | Home | 60637 | 606370000 | 170313202001 | 41.81030 | -87.59697 | 2020 |
| 101002 | 87654321 | 22334455 | 2024-11-04 15:00:00+00:00 UTC | 2024-11-07 11:00:00+00:00 UTC | 72 | Transfer from another hospital | Acute Care Transfer | Transferred to Acute Inpatient Rehab Facility | Acute Inpatient Rehab Facility | 46311 | 463110000 | 170313301002 | 41.55030 | -87.30101 | 2020 |
| 101003 | 11223344 | 11223344 | 2024-10-20 07:45:00+00:00 UTC | 2024-10-22 10:20:00+00:00 UTC | 59 | Pre-op surgical | Pre-op | Expired | Expired | 60446 | 604460000 | 170313401003 | 41.70010 | -87.60315 | 2020 |
labs Beta
The labs table is a long form (one lab result per row) longitudinal table.
| Column | Type | Description | Permissible Values |
|---|---|---|---|
| hospitalization_id | VARCHAR | ID variable for each patient encounter | No restriction |
| lab_order_dttm | DATETIME | Date and time when the lab is ordered. All datetime variables must be timezone-aware and set to UTC. | Datetime format should be YYYY-MM-DD HH:MM:SS+00:00 (UTC) |
| lab_collect_dttm | DATETIME | Date and time when the specimen is collected. All datetime variables must be timezone-aware and set to UTC. | Datetime format should be YYYY-MM-DD HH:MM:SS+00:00 (UTC) |
| lab_result_dttm | DATETIME | Date and time when the lab results are available. All datetime variables must be timezone-aware and set to UTC. | Datetime format should be YYYY-MM-DD HH:MM:SS+00:00 (UTC) |
| lab_order_name | VARCHAR | Procedure name for the lab, e.g. “Complete blood count w/ diff” | No restriction |
| lab_order_category | VARCHAR | Maps lab_order_name to standardized list of common lab order names, e.g. “CBC” | List of lab order categories in CLIF |
| lab_name | VARCHAR | Original lab component as recorded in the raw data, e.g. “AST (SGOT)”. | No restriction |
| lab_category | VARCHAR | Maps lab_name to a minimum set of standardized labs identified by the CLIF consortium as minimum necessary labs for the study of critical illness. | List of lab categories in CLIF |
| lab_value | VARCHAR | Recorded value corresponding to a lab. Lab values are often strings that can contain non-numeric results (e.g. “> upper limit of detection”). | No restriction |
| lab_value_numeric | DOUBLE | Parse out numeric part of the lab_value variable (optional). | Numeric |
| reference_unit | VARCHAR | Unit of measurement for that lab. | Permissible reference values for each lab_category listed here |
| lab_specimen_name | VARCHAR | Original fluid or tissue name the lab was collected from as given in the source data | No restriction |
| lab_specimen_category | VARCHAR | Fluid or tissue the lab was collected from, analogous to the LOINC “system” component. | working CDE c(blood/plasma/serum, urine, csf, other) |
| lab_loinc_code | VARCHAR | LOINC code for the lab | No restrictions |
| loinc_version | VARCHAR | Version or release of the LOINC coding system used for lab_loinc_code, e.g. 1.0 or 1.1. | No restriction |
Notes:
- All lab values must be reported using the lab’s reference units linked above in permissible values. Only the listed reference units are permissible for respective lab categories in the CLIF labs table. Sites must ensure that any raw laboratory values are converted to the reference units during the ETL process. Entries with other units must be transformed prior to loading into CLIF.
- The
lab_valuefield often has non-numeric entries that are useful to make project-specific decisions. A site may choose to keep thelab_valuefield as a character and create a new fieldlab_value_numericthat only parses the character field to extract the numeric part of the string.
Example:
| hospitalization_id | lab_order_dttm | lab_collect_dttm | lab_result_dttm | lab_order_name | lab_order_category | lab_name | lab_category | lab_value | lab_value_numeric | reference_unit | lab_specimen_name | lab_specimen_category | lab_loinc_code | loinc_version |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1001014 | 2023-05-01 07:00:00+00:00 UTC | 2023-05-01 07:15:00+00:00 UTC | 2023-05-01 08:00:00+00:00 UTC | Complete blood count w/ diff | CBC | WBC | white_blood_cell_count | 8.2 | 8.2 | 10^3/uL | blood | blood/plasma/serum | 6690-2 | 1.0 |
| 1001014 | 2023-05-01 07:00:00+00:00 UTC | 2023-05-01 07:15:00+00:00 UTC | 2023-05-01 08:00:00+00:00 UTC | Complete blood count w/ diff | CBC | HGB | hemoglobin | 13.5 | 13.5 | g/dL | blood | blood/plasma/serum | 718-7 | 1.0 |
| 1002025 | 2023-06-10 08:30:00+00:00 UTC | 2023-06-10 08:45:00+00:00 UTC | 2023-06-10 09:00:00+00:00 UTC | Basic metabolic panel | BMP | Sodium | sodium | 140 | 140 | mmol/L | blood | blood/plasma/serum | 2951-2 | 1.0 |
| 1002025 | 2023-06-10 08:30:00+00:00 UTC | 2023-06-10 08:45:00+00:00 UTC | 2023-06-10 09:00:00+00:00 UTC | Basic metabolic panel | BMP | Potassium | potassium | 4.2 | 4.2 | mmol/L | blood | blood/plasma/serum | 2823-3 | 1.0 |
| 1003036 | 2023-07-15 06:45:00+00:00 UTC | 2023-07-15 07:00:00+00:00 UTC | 2023-07-15 07:30:00+00:00 UTC | Liver function panel | LFT | AST (SGOT) | ast | 35 | 35 | U/L | blood | blood/plasma/serum | 1920-8 | 1.0 |
| 1003036 | 2023-07-15 06:45:00+00:00 UTC | 2023-07-15 07:00:00+00:00 UTC | 2023-07-15 07:30:00+00:00 UTC | Liver function panel | LFT | ALT (SGPT) | alt | 28 | 28 | U/L | blood | blood/plasma/serum | 1742-6 | 1.0 |
medication admin continuous Beta
This table captures medications administered at a rate over time, with NO set dose to be given. Examples include vasopressors, sedation, and paralysis drips. Multiple observations capture how the medication rate varies over time.
The medication admin continuous table is a long-form (one medication administration record per) longitudinal table designed for continuous infusions of common ICU medications such as vasopressors and sedation (Boluses of these drugs should be recorded in med_admin_intermittent). Note that it only reflects dose changes of the continuous medication and does not have a specific “end_time” variable to indicate the medication being stopped. The end of a continuous infusion should be recorded as a new row with med_dose = 0 and an appropriate mar_action_name (e.g. “stopped” or “paused”).
| Column | Type | Description | Permissible Values |
|---|---|---|---|
| hospitalization_id | VARCHAR | ID variable for each patient encounter | No restriction |
| med_order_id | VARCHAR | Medication order ID. Foreign key to link this table to other medication tables | No restriction |
| admin_dttm | DATETIME | Date and time when the medicine was administered. All datetime variables must be timezone-aware and set to UTC. | Datetime format should be YYYY-MM-DD HH:MM:SS+00:00 (UTC) |
| med_name | VARCHAR | Original med name string recorded in the raw data which often contains concentration e.g. NOREPInephrine 8 mg/250 mL | No restriction |
| med_category | VARCHAR | Maps med_name to a limited set of active ingredients for important ICU medications, e.g. norepinephrine | List of continuous medication categories in CLIF |
| med_group | VARCHAR | Limited number of ICU medication groups identified by the CLIF consortium, e.g. vasoactives | List of continuous medication groups in CLIF |
| med_route_name | VARCHAR | Medicine delivery route | e.g. IV, enteral |
| med_route_category | VARCHAR | Maps med_route_name to a standardized list of medication delivery routes. Refer to notes. | List of continuous route categories in CLIF |
| med_dose | FLOAT | Quantity of active drug delivered per unit time | Numeric |
| med_dose_unit | VARCHAR | Unit of dose in the format [active ingredient quantity]/[time] (e.g., mcg/min, mg/hr, units/hr, mcg/kg/min). Units are not standardized across drugs, and can be weight-based (e.g., mcg/kg/min). Boluses should be mapped to med_admin_intermittent | No restriction |
| infusion_rate | FLOAT | Absolute amount of volume administered over time, measured in [volume]/[time] (e.g., mL/hr). In contrast to med_dose which measures [quantity of drug]/[time] or [quantity of drug]/([time]*[body_weight]) | Numeric |
| infusion_rate_units | VARCHAR | Unit of infusion rate in the format [volume]/[time] (e.g., mL/hr, mL/min) | No restriction |
| mar_action_name | VARCHAR | MAR (medication administration record) action, e.g. stopped | No restriction |
| mar_action_category | VARCHAR | Maps mar_action_name to a standardized list of MAR actions | List of continuous action categories in CLIF |
| mar_action_group | VARCHAR | Maps mar_action_category to whether the action means the medication was administered or not. | administered, not_administered, other |
Example:
| hospitalization_id | admin_dttm | med_name | med_category | med_group | med_route_name | med_route_category | med_dose | med_dose_unit | infusion_rate | infusion_rate_units | mar_action_name | mar_action_group |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 792391 | 2123-11-13 12:28:00+00:00 UTC | PROPOFOL 10 MG/ML INTRAVENOUS EMULSION | propofol | sedation | Intravenous | NA | 75.0000 | mcg/kg/min | 45.0 | mL/hr | New Bag | administered |
| 792391 | 2123-11-13 13:49:00+00:00 UTC | REMIFENTANIL CONTINUOUS IV (ANESTHESIA) | remifentanil | sedation | NA | NA | 0.0500 | mcg/kg/min | 12.5 | mL/hr | New Bag | administered |
| 792391 | 2123-11-13 14:03:00+00:00 UTC | PROPOFOL 10 MG/ML INTRAVENOUS EMULSION | propofol | sedation | Intravenous | NA | 0.0000 | mcg/kg/min | 0.0 | mL/hr | Stopped | not_administered |
| 370921 | 2123-02-12 03:07:00+00:00 UTC | PHENYLEPHRINE 5 MG/50 ML (100 MCG/ML) IN 0.9 % SODIUM CHLORIDE | phenylephrine | vasoactives | Intravenous | NA | 20.0000 | mcg/min | 10.0 | mL/hr | New Bag | administered |
| 370921 | 2123-02-12 03:14:00+00:00 UTC | PHENYLEPHRINE 5 MG/50 ML (100 MCG/ML) IN 0.9 % SODIUM CHLORIDE | phenylephrine | vasoactives | Intravenous | NA | 50.0000 | mcg/min | 25.0 | mL/hr | Rate Change | administered |
| 702344 | 2123-04-27 04:30:00+00:00 UTC | HEPARIN (PORCINE) 25,000 UNIT/250 ML IN 0.45 % SODIUM CHLORIDE | heparin | anticoagulation | Intravenous | NA | 18.0000 | Units/kg/hr | 18.0 | mL/hr | New Bag | administered |
Notes:
- Include combination medications when mapping medication names to respective categories. Eg.
ACETAMIN-CALCIUM-MAG-CAFFEINE ORAL->acetaminophen - Include trial drugs when mapping medication names to respective categories… Eg.
ACETAMINOPHEN (IRB 140122) 325 MG ORAL TAB->acetaminophen
medication admin intermittent Beta
This table captures medications administered as fixed doses at discrete time points. Examples include antibiotics, steroids, and other medications given as boluses or scheduled doses. Each row represents ONE observation for each medication administered.
This table has exactly the same schema as medication_admin_continuous. The consortium decided to separate the medications that are administered intermittently from the continuously administered medications. The mCIDE for medication_category for intermittent meds can be found here.
| Column | Type | Description | Permissible Values |
|---|---|---|---|
| hospitalization_id | VARCHAR | ID variable for each patient encounter | No restriction |
| med_order_id | VARCHAR | Medication order ID. Foreign key to link this table to other medication tables | No restriction |
| admin_dttm | DATETIME | Date and time when the medicine was administered. All datetime variables must be timezone-aware and set to UTC. | Datetime format should be YYYY-MM-DD HH:MM:SS+00:00 (UTC) |
| med_name | VARCHAR | Original med name string recorded in the raw data which often contains concentration e.g. NOREPInephrine 8 mg/250 mL | No restriction |
| med_category | VARCHAR | Maps med_name to a limited set of active ingredients for important ICU medications, e.g. norepinephrine | List of medication categories in CLIF |
| med_group | VARCHAR | Limited number of ICU medication groups identified by the CLIF consortium, e.g. vasoactives | List of medication groups in CLIF |
| med_route_name | VARCHAR | Medicine delivery route | e.g. IV, enteral |
| med_route_category | VARCHAR | Maps med_route_name to a standardized list of medication delivery routes | List of intermittent route categories in CLIF |
| med_dose | FLOAT | Quantity taken in dose | Numeric |
| med_dose_unit | VARCHAR | Unit of dose. e.g. mcg. | No restriction |
| mar_action_name | VARCHAR | MAR (medication administration record) action, e.g. stopped | No restriction |
| mar_action_category | VARCHAR | Maps mar_action_name to a standardized list of MAR actions | List of intermittent action categories in CLIF |
| mar_action_group | VARCHAR | Maps mar_action_category to whether the action means the medication was administered or not. | administered, not_administered, other |
Notes:
- Continuous medications are included in this table when given as boluses
- Intermittent medications can be given at different rates
microbiology culture Beta
The microbiology culture table is a wide longitudinal table that captures the order and result times of microbiology culture tests, the type of fluid collected, the component of the test, and the organism identified.
| Column | Type | Description | Permissible Values |
|---|---|---|---|
| patient_id | VARCHAR | Unique identifier for each patient, presumed to be a distinct individual. | No restriction |
| hospitalization_id | VARCHAR | ID variable for each patient encounter | No restriction |
| organism_id | VARCHAR | Distinct numerical identifier that each site creates which links a unique, non-missing organism_category that has a distinct patient_id, hospitalization_id, lab_order_dttm, lab_collect_dttm, lab_result_dttm, and fluid_category with its method_category == culture from the microbiology_culture table to an antibiotic_category and susceptibility_category from the microbiology_susceptibility table | No restriction |
| order_dttm | DATETIME | Date and time when the test is ordered. | Datetime format should be YYYY-MM-DD HH:MM:SS+00:00 |
| collect_dttm | DATETIME | Date and time when the specimen is collected. | Datetime format should be YYYY-MM-DD HH:MM:SS+00:00 |
| result_dttm | DATETIME | Date and time when the results are available. | Datetime format should be YYYY-MM-DD HH:MM:SS+00:00 |
| fluid_name | VARCHAR | Fluid name string from the raw data. | No restriction. |
| fluid_category | VARCHAR | Fluid categories defined according to the NIH common data elements. | CDE NIH Infection Site |
| method_name | VARCHAR | Original method names from the source data. | No restriction |
| method_category | VARCHAR | Maps method_name to a standardized list of method categories. | culture, gram stain, smear |
| organism_name | VARCHAR | Organism name from the raw data. | No restriction. |
| organism_category | VARCHAR | Maps organism_name to the standardized list of organisms under the structure of genus species. | Organism species. Check this file for examples. |
| organism_group | VARCHAR | Maps organism_category to the standardized list of organisms under the NIH CDE structure. | CDE NIH Organism |
| lab_loinc_code | VARCHAR | LOINC code. | No restriction |
Example:
| patient_id | hospitalization_id | organism_id | order_dttm | collect_dttm | result_dttm | fluid_name | fluid_category | method_name | method_category | organism_name | organism_category | organism_group | lab_loinc_code |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 12345 | HOSP12345 | ORG001 | 2025-06-05 08:15:00+00:00 | 2025-06-05 08:45:00+00:00 | 2025-06-06 12:00:00+00:00 | AFB/FUNGAL BLOOD CULTURE | Blood/Buffy Coat | Blood culture | culture | Acinetobacter baumanii | acinetobacter_baumanii | acinetobacter (baumanii, calcoaceticus, lwoffi, other species) | |
| 12345 | HOSP12345 | ORG002 | 2025-06-05 08:15:00+00:00 | 2025-06-05 08:45:00+00:00 | 2025-06-06 12:00:00+00:00 | AFB/FUNGAL BLOOD CULTURE | Blood/Buffy Coat | Blood culture | culture | Candida albicans | candida_albicans | candida albicans | |
| 67890 | HOSP67890 | ORG003 | 2025-06-10 14:10:00+00:00 | 2025-06-10 14:35:00+00:00 | 2025-06-11 09:20:00+00:00 | BRAIN BIOPSY CULTURE | Brain | Tissue culture | culture | Aspergillus fumigatus | aspergillus_fumigatus | asperguillus fumigatus |
microbiology susceptibility Beta
This table is used to store the susceptibility results of the organisms identified in the Microbiology Culture.
| Column | Type | Description | Permissible Values |
|---|---|---|---|
| organism_id | VARCHAR | Distinct numerical identifier that each site creates which links a unique, non-missing organism_category that has a distinct patient_id, hospitalization_id, lab_order_dttm, lab_collect_dttm, lab_result_dttm, and fluid_category with its method_category == culture from the microbiology_culture table to an antibiotic_category and susceptibility_category from the microbiology_susceptibility table | No restriction |
| antimicrobial_name | VARCHAR | Name of the antimicrobial | No restriction |
| antimicrobial_category | VARCHAR | Category or class of the antimicrobial tested | List of antimicrobial categories in CLIF |
| sensitivity_name | VARCHAR | Name of the test result used to determine susceptibility (e.g., value of mcg/mL or MIC) | No restriction |
| susceptibility_name | VARCHAR | Name of the sensitivity interpretation | No restriction |
| susceptibility_category | VARCHAR | Standardized category of susceptibility. | susceptible, non susceptible, indeterminate, NA |
Example:
| organism_id | antimicrobial_name | antimicrobial_category | sensitivity_name | susceptibility_name | susceptibility_category |
|---|---|---|---|---|---|
| 1 | avycex (ceftazidime/avibactam) | ceftazidime_avibactam | 8 MIC | susceptible | susceptible |
| 1 | amoxicillin clavulanic acid | amoxicillin_clavulanate | < 0.1 ug/mL | susceptible | susceptible |
| 1 | meropenem | meropenem | indeterminate | indeterminate | indeterminate |
| 1 | ampicillin 500mg | ampicillin | > 0.5 ug/mL | resistant | NA |
| 2 | unasyn | ampicillin_sulbactam | susceptible dose dependent | susceptible | susceptible |
| 2 | Ertapenem | ertapenem | not reported | NA | NA |
| 2 | Vancomycin (non-Cdiff) | vancomycin | > 0.25 ug/mL | non susceptible, caution | non_susceptible |
patient Beta
This table contains demographic information about the patient that does not vary between hospitalizations. It is inspired by the OMOP Person table
| Column | Type | Description | Permissible Values |
|---|---|---|---|
| patient_id | VARCHAR | Unique identifier for each patient, presumed to be a distinct individual. | No restriction |
| race_name | VARCHAR | Patient race string from source data | No restriction |
| race_category | VARCHAR | A standardized CDE description of patient race per the US Census | Black or African American, White, American Indian or Alaska Native, Asian, Native Hawaiian or Other Pacific Islander, Unknown, Other |
| ethnicity_name | VARCHAR | Patient ethnicity string from source data | No restriction |
| ethnicity_category | VARCHAR | Description of patient ethnicity per the US census definition | Hispanic, Non-Hispanic, Unknown |
| sex_name | VARCHAR | Patient’s biological sex as given in the source data | No restriction |
| sex_category | VARCHAR | Patient biological sex | Male, Female, Unknown |
| birth_date | DATE | Patient’s date of birth | Date format should be YYYY-MM-DD |
| death_dttm | DATETIME | Patient’s death date, including time. Can be sourced from hospital records, or external sources like state vital records registries | Datetime format should be YYYY-MM-DD HH:MM:SS+00:00 (UTC) |
| language_name | VARCHAR | Patient’s preferred language | Original string from the source data |
| language_category | VARCHAR | Maps language_name to a standardized list of spoken languages | List of language categories in CLIF |
Example:
| patient_id | race_name | race_category | ethnicity_name | ethnicity_category | sex_category | birth_date | death_dttm | language_name | language_category |
|---|---|---|---|---|---|---|---|---|---|
| 132424 | Black or African-American | Black or African American | Not Hispanic, Latino/a, or Spanish origin | Non-Hispanic | Male | 2145-05-09 | NA | English | English |
| 132384 | White | White | Not Hispanic, Latino/a, or Spanish origin | Non-Hispanic | Female | 2145-03-30 | NA | English | English |
| 542367 | Black or African-American | Black or African American | Not Hispanic, Latino/a, or Spanish origin | Non-Hispanic | Male | 2145-01-29 | NA | English | English |
| 989862 | White | White | Not Hispanic, Latino/a, or Spanish origin | Non-Hispanic | Female | 2145-11-06 | NA | English | English |
| 428035 | More than one Race | Other | Not Hispanic, Latino/a, or Spanish origin | Non-Hispanic | Male | 2145-10-13 | NA | English | English |
patient assessments Beta
The patient_assessments table captures various assessments performed on patients across different domains, including neurological status, sedation levels, pain, and withdrawal. The table is designed to provide detailed information about the assessments, such as the name of the assessment, the category, and the recorded values.
| Column | Type | Description | Permissible Values |
|---|---|---|---|
| hospitalization_id | VARCHAR | ID variable for each patient encounter | No restriction |
| recorded_dttm | DATETIME | The exact date and time when the assessment was recorded, ensuring temporal accuracy. All datetime variables must be timezone-aware and set to UTC. | Datetime format should be YYYY-MM-DD HH:MM:SS+00:00 (UTC) |
| assessment_name | VARCHAR | Assessment Tool Name. The primary name of the assessment tool used (e.g., GCS, NRS, SAT Screen). | No restriction |
| assessment_category | VARCHAR | Maps assessment_name to a standardized list of patient assessments | List of permissible assessment categories in CLIF |
| assessment_group | VARCHAR | Broader Assessment Group. This groups the assessments into categories such as Sedation, Neurologic, Pain, etc. | List of permissible assessment groups in CLIF |
| numerical_value | DOUBLE | Numerical Assessment Result. The numerical result or score from the assessment component. | Applicable for assessments with numerical outcomes (e.g., 0-10, 3-15) |
| categorical_value | VARCHAR | Categorical Assessment Result. The categorical outcome from the assessment component. | Applicable for assessments with categorical outcomes (e.g., Pass/Fail, Yes/No) |
| text_value | VARCHAR | Textual Assessment Result. The textual explanation or notes from the assessment component. | Applicable for assessments requiring textual data |
Example:
| hospitalization_id | recorded_dttm | assessment_name | assessment_category | assessment_group | numerical_value | categorical_value | text_value |
|---|---|---|---|---|---|---|---|
| 12345 | 2024-12-01 08:15:00+00:00 UTC | NUR RA GLASGOW ADULT EYE OPENING | gcs_eye | Neurological | 4 | NA | NA |
| 12345 | 2024-12-01 08:15:00+00:00 UTC | NUR RA GLASGOW ADULT VERBAL RESPONSE | gcs_verbal | Neurological | 5 | NA | NA |
| 12345 | 2024-12-01 08:15:00+00:00 UTC | NUR RA GLASGOW ADULT BEST MOTOR RESPONSE | gcs_motor | Neurological | 6 | NA | NA |
| 12345 | 2024-12-01 08:15:00+00:00 UTC | NUR RA GLASGOW ADULT SCORING | gcs_total | Neurological | 15 | NA | NA |
| 67890 | 2024-12-01 10:30:00+00:00 UTC | BRADEN ASSESSMENT | braden_total | Nursing Risk | 18 | NA | NA |
| 67890 | 2024-12-01 10:30:00+00:00 UTC | SAT SCREEN | sat_delivery_pass_fail | Sedation | NA | Pass | NA |
patient procedures Beta
A long table of standardized procedural billing codes associated with the hospitalization, using the Healthcare Common Procedure Coding System (HCPCS). In CLIF version 2.1, the patient_procedures table includes only procedures that were actually performed or completed (not cancelled), and only contains professional billing codes—specifically, CPT codes billed by clinicians (HCPCS Level 1).
Hospital billing i.e., Products, supplies, and services that do not have CPT codes (HCPCS Level 2) are not included in this table.
Additionally, this table contains ICD-10-PCS procedure codes which are not used for clinician billing but can contribute to the calculation of DRGs for hospital reimbursement and can also appear in the hospital_diagnosis table.
| Column | Type | Description | Permissible Values |
|---|---|---|---|
| hospitalization_id | VARCHAR | ID variable for each patient encounter | No restriction |
| billing_provider_id | VARCHAR | Uniquely identifies the billingprovider associated with the procedure. | No restriction |
| performing_provider_id | VARCHAR | Uniquely identifies the performing provider associated with the procedure. | No restriction |
| procedure_code | VARCHAR | Encoded procedure identifier. | Valid CPT, ICD-10-PCS OR HCPCS code |
| procedure_code_format | VARCHAR | Code format used. | CPT, ICD10PCS, HCPCS |
| procedure_billed_dttm | DATETIME | Date and time the procedure was billed (may differ from actual procedure time). All datetime variables must be timezone-aware and set to UTC. | Datetime format should be YYYY-MM-DD HH:MM:SS+00:00 (UTC) |
Example:
| hospitalization_id | billing_provider_id | performing_provider_id | procedure_code | procedure_code_format | procedure_billed_dttm |
|---|---|---|---|---|---|
| HOSP1001 | BP123 | PP456 | 36556 | CPT | 2024-01-01 08:00:00+00:00 UTC |
| HOSP1001 | BP123 | PP789 | 32551 | CPT | 2024-01-01 10:00:00+00:00 UTC |
| HOSP1002 | BP234 | PP890 | G0009 | HCPCS | 2024-01-05 09:30:00+00:00 UTC |
| HOSP1002 | BP234 | PP890 | G0008 | HCPCS | 2024-01-05 11:00:00+00:00 UTC |
| HOSP1003 | BP345 | PP901 | 36620 | CPT | 2024-01-10 07:00:00+00:00 UTC |
position Beta
The position table is a long form (one position per row) longitudinal table that captures all documented position changes of the patient. The table is designed for the explicit purpose of constructing the position_category CDE and identifying patients in prone position.
| Column | Type | Description | Permissible Values |
|---|---|---|---|
| hospitalization_id | VARCHAR | ID variable for each patient encounter | No restriction |
| recorded_dttm | DATETIME | Date and time when the vital is recorded. All datetime variables must be timezone-aware and set to UTC. | Datetime format should be YYYY-MM-DD HH:MM:SS+00:00 |
| position_name | VARCHAR | Description of the position from the source data. This field is not used for analysis. | No restriction |
| position_category | VARCHAR | Maps position_name to either prone or not prone. | prone, not_prone |
Example:
| hospitalization_id | recorded_dttm | position_name | position_category |
|---|---|---|---|
| 84 | 2123-06-20 00:00:00+00:00 UTC | Supine–turn R | not_prone |
| 84 | 2123-06-20 06:00:00+00:00 UTC | Supine–turn L | not_prone |
| 84 | 2123-06-20 12:00:00+00:00 UTC | Supine–back | not_prone |
| 84 | 2123-06-20 16:00:00+00:00 UTC | Supine–turn R | not_prone |
| 84 | 2123-06-20 20:00:00+00:00 UTC | Supine–back;Supine–turn intolerant | not_prone |
| 84 | 2123-06-20 22:00:00+00:00 UTC | Supine–turn intolerant,microturn L | not_prone |
| 84 | 2123-06-20 00:00:00+00:00 UTC | Supine–turn intolerant,microturn L;Supine–back | not_prone |
| 84 | 2123-06-20 01:10:00+00:00 UTC | 30 Degrees | not_prone |
respiratory support Beta
The respiratory support table is a wider longitudinal table that captures simultaneously recorded ventilator settings and observed ventilator parameters. The table is designed to capture the most common respiratory support devices and modes used in the ICU. It will be sparse for patients who are not on mechanical ventilation.
| Column | Type | Description | Permissible Values |
|---|---|---|---|
| hospitalization_id | VARCHAR | ID variable for each patient encounter | No restriction |
| recorded_dttm | DATETIME | Date and time when the device settings and/or measurement was recorded. All datetime variables must be timezone-aware and set to UTC. | Datetime format should be YYYY-MM-DD HH:MM:SS+00:00 |
| device_name | VARCHAR | Raw string of the device. | No restriction |
| device_id | VARCHAR | Unique ID of the individual physical device used (e.g. ventilator ACZ3RV91), enables linkage to continuous waveform data. Distinct from vent_brand_name, which stores the brand or model. | No restriction |
| device_category | VARCHAR | Maps device_name to a standardized list of respiratory support device categories | IMV, NIPPV, CPAP, High Flow NC, Face Mask, Trach Collar, Nasal Cannula, T Piece, Room Air, Other |
| vent_brand_name | VARCHAR | Ventilator model name when device_category is IMV or NIPPV | Optional |
| mode_name | VARCHAR | Raw string of ventilation mode (e.g., CMV volume control) | No restriction |
| mode_category | VARCHAR | Standardized list of modes of mechanical ventilation | Assist Control-Volume Control, Pressure Control, Pressure-Regulated Volume Control, SIMV, Pressure Support/CPAP, Volume Support, Blow by, Other |
| tracheostomy | INT | Indicates if tracheostomy is present | 0 = No, 1 = Yes |
| fio2_set | FLOAT | Fraction of inspired oxygen set (e.g., 0.21) | No restriction, see Expected _set values for each device_category and mode_category |
| lpm_set | FLOAT | Liters per minute of supplemental oxygen set for patients NOT on positive pressure ventilation | No restriction, see Expected _set values for each device_category and mode_category |
| tidal_volume_set | FLOAT | Tidal volume set (in mL) | No restriction, see Expected _set values for each device_category and mode_category |
| resp_rate_set | FLOAT | Respiratory rate set (in bpm) | No restriction, see Expected _set values for each device_category and mode_category |
| pressure_control_set | FLOAT | Pressure control set (in cmH2O) | No restriction, see Expected _set values for each device_category and mode_category |
| pressure_support_set | FLOAT | Pressure support set (in cmH2O) | No restriction, see Expected _set values for each device_category and mode_category |
| flow_rate_set | FLOAT | Flow rate of air delivered to patients on non-invasive devives (in lpm) | No restriction, see Expected _set values for each device_category and mode_category |
| peak_inspiratory_pressure_set | FLOAT | Peak inspiratory pressure set (in cmH2O). This is equivalent to inspiratory positive airway pressure (IPAP) in non-invasive ventilation modes, and is not used in invasive ventilation modes. | No restriction, see Expected _set values for each device_category and mode_category |
| inspiratory_time_set | FLOAT | Inspiratory time set (in seconds) | No restriction, see Expected _set values for each device_category and mode_category |
| peep_set | FLOAT | Positive-end-expiratory pressure set (in cmH2O) | No restriction, see Expected _set values for each device_category and mode_category |
| tidal_volume_obs | FLOAT | Observed tidal volume (in mL) | No restriction |
| resp_rate_obs | FLOAT | Observed respiratory rate (in bpm), as measured and recorded by the ventilator or non-invasive device. This value should not be pulled from the general vitals table, but should reflect measurements from the respiratory support device itself. | No restriction |
| plateau_pressure_obs | FLOAT | Observed plateau pressure (in cmH2O) | No restriction |
| peak_inspiratory_pressure_obs | FLOAT | Observed peak inspiratory pressure (in cmH2O) | No restriction |
| peep_obs | FLOAT | Observed PEEP (in cmH2O) | No restriction |
| minute_vent_obs | FLOAT | Observed minute ventilation (in liters) | No restriction |
| mean_airway_pressure_obs | FLOAT | Observed mean airway pressure | No restriction |
Notes:
Expected setting values for each device_category and mode_category
device_category== “IMV”
| ventilator_setting | Assist Control-Volume Control | Pressure Support/CPAP | Pressure Control | Pressure-Regulated Volume Control | SIMV | Volume Support |
|---|---|---|---|---|---|---|
| fio2_set | E | E | E | E | E | E |
| tidal_volume_set | E | E | P | E | ||
| resp_rate_set | E | E | E | E | ||
| pressure_control_set | E | P | ||||
| pressure_support_set | E | E | ||||
| flow_rate_set | P | P | ||||
| inspiratory_time_set | P | E | P | |||
| peep_set | E | E | E | E | E | E |
E = Expected ventilator setting for the mode, P = possible ventilator setting for the mode.
device_category== “NIPPV”
mode_category is Pressure Support/CPAP and the fio2_set, peep_set, and either pressure_support_set OR peak_inspiratory_pressure_set (IPAP) is required.
device_category== “CPAP”
mode_category is Pressure Support/CPAP and the fio2_set and peep_set are required.
device_category== “High Flow NC”
mode_category is NA and the fio2_set and lpm_set are required.
device_category== “Face Mask”
mode_category is NAlpm_set is required.fio2_set is possible.
device_category== “Trach Collar” or “Nasal Cannula”
mode_category is NAlpm_set is required
Example:
| hospitalization_id | recorded_dttm | device_name | device_id | device_category | mode_name | mode_category | vent_brand_name | tracheostomy | fio2_set | lpm_set | tidal_volume_set | resp_rate_set | pressure_control_set | pressure_support_set | flow_rate_set | tidal_volume_obs | resp_rate_obs | plateau_pressure_obs | peak_inspiratory_pressure_obs | peep_obs | minute_vent_obs | mean_airway_pressure_obs |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 12345 | 2024-12-01 08:00:00+00:00 UTC | Ventilator | DEV001 | IMV | CMV Volume Ctrl | Assist Control-Volume Control | Vent A | 1 | 0.50 | 40 | 500 | 18 | 15 | 5 | 50 | 450 | 18 | 20 | 25 | 5 | 9.0 | 12.0 |
| 12345 | 2024-12-01 09:00:00+00:00 UTC | Ventilator | DEV001 | IMV | SIMV | SIMV | Vent A | 1 | 0.45 | 35 | 480 | 20 | 18 | 8 | 55 | 470 | 20 | 21 | 28 | 6 | 10.5 | 14.0 |
| 67890 | 2024-12-01 10:30:00+00:00 UTC | HFNC | DEV002 | High Flow NC | N/A | Other | N/A | 0 | 0.30 | 60 | NA | NA | NA | NA | 60 | NA | NA | NA | NA | NA | NA | NA |
| 67890 | 2024-12-01 11:00:00+00:00 UTC | CPAP | DEV003 | CPAP | CPAP | Pressure Support/CPAP | CPAP X | 0 | 0.40 | 50 | NA | NA | NA | 10 | NA | NA | NA | NA | NA | 8 | NA | NA |
vitals Beta
The vitals table is a long-form (one vital sign per row) longitudinal table.
| Column | Type | Description | Permissible Values |
|---|---|---|---|
| hospitalization_id | VARCHAR | ID variable for each patient encounter | No restriction |
| recorded_dttm | DATETIME | Date and time when the vital is recorded. All datetime variables must be timezone-aware and set to UTC. | Datetime format should be YYYY-MM-DD HH:MM:SS+00:00 (UTC) |
| vital_name | VARCHAR | Description of the flowsheet measure from the source data. Not used for analysis. | No restriction |
| vital_category | VARCHAR | Maps vital_name to a list of standard vital sign categories. | temp_c, heart_rate, sbp, dbp, spo2, respiratory_rate, map, height_cm, weight_kg |
| vital_value | FLOAT | Recorded value of the vital. Measurement unit should match the vital category. | temp_c = Celsius, height_cm = Centimeters, weight_kg = Kg, map = mmHg, spo2 = %. No unit for heart_rate, sbp, dbp, respiratory_rate |
| meas_site_name | VARCHAR | Site where the vital is recorded. Optional field with no associated category. | No restriction |
Example:
| hospitalization_id | recorded_dttm | vital_name | vital_category | vital_value | meas_site_name |
|---|---|---|---|---|---|
| 20010012 | 2024-12-01 08:00:00+00:00 UTC | HEIGHT | height_cm | 170.0 | unspecified |
| 20010012 | 2024-12-01 08:15:00+00:00 UTC | WEIGHT | weight_kg | 70.0 | unspecified |
| 20010012 | 2024-12-01 08:30:00+00:00 UTC | PULSE | heart_rate | 72.0 | unspecified |
| 20010012 | 2024-12-01 08:45:00+00:00 UTC | BLOOD PRESSURE (SYSTOLIC) | sbp | 120.0 | unspecified |
| 20010012 | 2024-12-01 08:45:00+00:00 UTC | BLOOD PRESSURE (DIASTOLIC) | dbp | 80.0 | unspecified |
| 20010012 | 2024-12-01 08:50:00+00:00 UTC | RESPIRATORY RATE | respiratory_rate | 16.0 | unspecified |
| 20010012 | 2024-12-01 09:00:00+00:00 UTC | TEMPERATURE | temp_c | 36.8 | unspecified |
| 20010012 | 2024-12-01 09:15:00+00:00 UTC | SPO2 | spo2 | 98.0 | unspecified |
| 20010013 | 2024-12-01 09:30:00+00:00 UTC | MEAN ARTERIAL PRESSURE (MAP) | map | 85.0 | arterial |
Concept Tables
A planned future CLIF table that has yet to be used in a federated project. The table structure and CDE elements are in draft form. Permissible values of category variables may still need to be defined. Seeking conceptual feedback. Significant changes to all aspects of the table are possible.
clinical trial Concept
This table captures whether a patient was enrolled in any clinical trial during their hospitalization. It enables longitudinal tracking of trial participation, including trial identifiers, arm assignment, and key consent, randomization, and withdrawal timestamps. This structure supports research into the effects of experimental therapies and interventions on patient outcomes.
| Column | Type | Description | Permissible Values |
|---|---|---|---|
| participant_id | VARCHAR | Unique identifier for each clinical trial participant from clinical trial management software, used for pipelines to electronic data capture software. NOT the same as patient_id | No restriction |
| patient_id | VARCHAR | Unique identifier for each patient, presumed to be a distinct individual. | No restriction |
| hospitalization_id | VARCHAR | ID variable for each patient encounter. | No restriction |
| trial_id | VARCHAR | Unique identifier for the clinical trial (e.g., institutional trial ID, NCT number). | No restriction |
| trial_name | VARCHAR | Descriptive name of the clinical trial. | No restriction |
| arm_id | VARCHAR | Identifier indicating which arm of the trial the participant was enrolled in. | No restriction |
| consent_dttm | DATETIME | Timestamp of participant consent for the clinical trial. All datetime variables must be timezone-aware and set to UTC. | Datetime format should be YYYY-MM-DD HH:MM:SS+00:00 (UTC) |
| enrollment_dttm | DATETIME | Timestamp of participant enrollment in the clinical trial. All datetime variables must be timezone-aware and set to UTC. | Datetime format should be YYYY-MM-DD HH:MM:SS+00:00 (UTC) |
| randomized_dttm | DATETIME | Timestamp of participant randomization in the clinical trial. All datetime variables must be timezone-aware and set to UTC. | Datetime format should be YYYY-MM-DD HH:MM:SS+00:00 (UTC) |
| withdrawal_dttm | DATETIME | Timestamp of trial withdrawal if applicable. All datetime variables must be timezone-aware and set to UTC. | Datetime format should be YYYY-MM-DD HH:MM:SS+00:00 (UTC) |
Example:
| participant_id | patient_id | hospitalization_id | trial_id | trial_name | arm_id | consent_dttm | enrollment_dttm | randomized_dttm | withdrawal_dttm |
|---|---|---|---|---|---|---|---|---|---|
| PT1234 | 567890 | 234567 | T-001 | Early Vasopressor in Septic Shock (EVSS) | Arm A | 2024-06-01 10:15:00+00:00 UTC | 2024-06-01 14:00:00+00:00 UTC | 2024-06-01 15:00:00+00:00 UTC | |
| PT2345 | 678901 | 345678 | NCT04321 | Lung Protective Ventilation | Standard | 2024-06-11 12:00:00+00:00 UTC | 2024-06-11 13:15:00+00:00 UTC | ||
| PT3456 | 789012 | 456789 | T-017 | Multi-center COVID-19 Anticoagulation Trial | High Dose | 2024-07-15 09:10:00+00:00 UTC | 2024-07-15 09:45:00+00:00 UTC | 2024-07-15 10:00:00+00:00 UTC | 2024-07-20 08:30:00+00:00 UTC |
ecmo mcs Concept
The ECMO/MCS table is a wider longitudinal table that captures the start and stop times of ECMO/MCS support, the type of device used, and the work rate of the device.
| Column | Type | Description | Permissible Values |
|---|---|---|---|
| hospitalization_id | VARCHAR | ID variable for each patient encounter | No restriction |
| recorded_dttm | DATETIME | Date and time when the device settings and/or measurement was recorded | Datetime format should be YYYY-MM-DD HH:MM:SS+00:00 (UTC) |
| device_name | VARCHAR | Name of the ECMO/MCS device used including brand information, e.g. Centrimag | No restriction |
| device_category | VARCHAR | Maps device_name to a standardized mCIDE | List of device categories in CLIF and outlier thresholds by device category |
| mcs_group | VARCHAR | Maps device_category to a standardized mCIDE of MCS types | List of MCS groups in CLIF |
| ecmo_configuration_category | VARCHAR | Categorical variable designating the ECMO configuration type as defined by the cannulation strategy | vv, va, va_v, vv_a, etc. |
| control_parameter_name | VARCHAR | String that captures the measure of work rate of the device, e.g., RPMs, Impella Power, etc. | No restriction |
| control_parameter_category | VARCHAR | Maps control_parameter_name to a standardized list of control parameter categories | List of control parameter categories in CLIF |
| control_parameter_value | FLOAT | The value of the control parameter (numeric). | Numeric values |
| flow | FLOAT | Blood flow in L/min. | Numeric values in L/min |
| sweep_set | FLOAT | Gas flow (L/min) set. Applies to ECMO only. | Numeric values in L/min |
| fdO2_set | FLOAT | Fraction of delivered oxygen set. Applies to ECMO only. | Numeric values (0-1) |
Example:
| hospitalization_id | recorded_dttm | device_name | device_category | mcs_group | ecmo_configuration_category | control_parameter_name | control_parameter_category | control_parameter_value | flow | sweep_set | fdO2_set |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 21002 | 2024-02-18 09:10:00+00:00 UTC | Impella 5.5 | impella_5_5 | impella_lvad | NULL | P5 | impella_performance | 5 | 4.8 | NULL | NULL |
| 21003 | 2024-02-22 11:40:00+00:00 UTC | Abbott IABP | iabp | iabp | NULL | IABP Ratio | iabp_ratio | 3 | NULL | NULL | NULL |
| 21004 | 2024-02-23 14:30:00+00:00 UTC | Heart Mate III LVAD | heartmate_3 | durable_lvad | NULL | RPMs | rpm | 4200 | 3.5 | NULL | NULL |
| 21005 | 2024-02-27 16:25:00+00:00 UTC | Centrimag ECMO VV | centrimag_ecmo | ecmo | vv | RPMs | rpm | 3500 | 3 | 1 | NULL |
intake output Concept
The intake_output table is long form table that captures the times intake and output events were recorded, the type of fluid administered or recorded as “out”, and the amount of fluid.
| Column | Type | Description | Permissible Values |
|---|---|---|---|
| hospitalization_id | VARCHAR | ID variable for each patient encounter | No restriction |
| intake_dttm | DATETIME | Date and time of intake. All datetime variables must be timezone-aware and set to UTC. | Datetime format should be YYYY-MM-DD HH:MM:SS+00:00 (UTC) |
| fluid_name | VARCHAR | Name of the fluid administered. | No restriction |
| amount | DOUBLE | Amount of fluid administered (in mL) | Numeric values in mL |
| in_out_flag | INT | Indicator for intake or output (1 for intake, 0 for output) | 0 = Output, 1 = Intake |
Example:
| hospitalization_id | intake_dttm | fluid_name | amount | in_out_flag |
|---|---|---|---|---|
| 1001 | 2024-01-01 08:00:00+00:00 UTC | Normal Saline | 500 | 1 |
| 1001 | 2024-01-01 10:30:00+00:00 UTC | Urine | 300 | 0 |
| 1002 | 2024-01-05 09:15:00+00:00 UTC | Dextrose | 250 | 1 |
| 1002 | 2024-01-05 14:00:00+00:00 UTC | Urine | 400 | 0 |
| 1003 | 2024-01-10 07:45:00+00:00 UTC | Lactated Ringer’s | 600 | 1 |
| 1003 | 2024-01-10 12:00:00+00:00 UTC | Drainage | 200 | 0 |
invasive hemodynamics Concept
The invasive_hemodynamics table records invasive hemodynamic measurements during a patient’s hospitalization. These measurements represent pressures recorded via invasive monitoring and are expressed in millimeters of mercury (mmHg).
| Column | Type | Description | Permissible Values |
|---|---|---|---|
| hospitalization_id | VARCHAR | ID variable for each patient encounter | No restriction |
| recorded_dttm | DATETIME | The date and time when the measurement was recorded. All datetime variables must be timezone-aware and set to UTC. | Datetime format should be YYYY-MM-DD HH:MM:SS+00:00 (UTC) |
| measure_name | VARCHAR | Description of the site or context of the invasive hemodynamic measurement. | Free text (e.g., Right Atrium) |
| measure_category | VARCHAR | Categorical variable specifying the type of invasive hemodynamic measurement. | cvp, ra, rv, pa_systolic, pa_diastolic, pa_mean, pcwp, cardiac_output_thermodilution , cardiac_output_fick |
| measure_value | DOUBLE | The numerical value of the invasive hemodynamic measurement in mmHg. | Positive decimal values (e.g., 5.00, 25.65) |
Notes:
- All
measure_valueentries should be recorded in mmHg. - The
measure_categoryfield ensures standardization of invasive hemodynamic data:
1.CVP- Central Venous Pressure
2.RA- Right Atrial Pressure
3.RV- Right Ventricular Pressure
4.PA_systolic- Pulmonary Artery Systolic Pressure
5.PA_diastolic- Pulmonary Artery Diastolic Pressure
6.PA_mean- Pulmonary Artery Mean Pressure
7.PCWP- Pulmonary Capillary Wedge Pressure
Example:
| hospitalization_id | recorded_dttm | measure_name | measure_category | measure_value |
|---|---|---|---|---|
| 12345 | 2024-12-01 08:30:00+00:00 UTC | CVP | CVP | 12.50 |
| 12345 | 2024-12-01 09:00:00+00:00 UTC | Pulmonary Artery-Sys | PA_systolic | 25.00 |
| 12345 | 2024-12-01 09:30:00+00:00 UTC | Wedge | PCWP | 18.75 |
key icu orders Concept
The key_icu_orders table captures key orders related to physical therapy (PT) and occupational therapy (OT) during ICU stays. It includes details about the hospitalization, the timing of the order, the specific name of the order, its category, and the status of the order (completed or sent).
| Column | Type | Description | Permissible Values |
|---|---|---|---|
| hospitalization_id | VARCHAR | ID variable for each patient encounter | No restriction |
| order_dttm | DATETIME | Date and time when the order was placed. All datetime variables must be timezone-aware and set to UTC. | Datetime format should be YYYY-MM-DD HH:MM:SS+00:00 (UTC) |
| order_name | VARCHAR | Name of the specific order (e.g., PT Evaluation, OT Treatment). | No restriction |
| order_category | VARCHAR | Category of the order. | Under-development. Some examples include: pt_evaluation, pt_treat, ot_evaluation, ot_treat |
| order_status_name | VARCHAR | Status of the order. | sent, completed |
Example:
| hospitalization_id | order_dttm | order_name | order_category | order_status_name |
|---|---|---|---|---|
| 12345 | 2024-12-15 10:00:00+00:00 UTC | PT Initial Evaluation | PT_evaluation | completed |
| 67890 | 2024-12-16 14:30:00+00:00 UTC | OT Follow-up Treatment | OT_treat | sent |
| 54321 | 2024-12-16 08:00:00+00:00 UTC | PT Mobility Session | PT_treat | completed |
| 98765 | 2024-12-15 11:15:00+00:00 UTC | OT Cognitive Assessment | OT_evaluation | sent |
medication orders Concept
This table records the ordering (not administration) of medications. The table is in long form (one medication order per row) longitudinal table. Linkage to the medication_admin_continuous and medication_admin_intermittent tables is through the med_order_id field.
| Column | Type | Description | Permissible Values |
|---|---|---|---|
| hospitalization_id | VARCHAR | ID variable for each patient encounter | No restriction |
| med_order_id | VARCHAR | Unique identifier for each medication order | No restriction |
| order_start_dttm | DATETIME | Date and time when the medication order was initiated. All datetime variables must be timezone-aware and set to UTC. | Datetime format should be YYYY-MM-DD HH:MM:SS+00:00 (UTC) |
| order_end_dttm | DATETIME | Date and time when the medication order ended or was discontinued. All datetime variables must be timezone-aware and set to UTC. | Datetime format should be YYYY-MM-DD HH:MM:SS+00:00 (UTC) |
| ordered_dttm | DATETIME | Date and time when the medication was actually ordered. All datetime variables must be timezone-aware and set to UTC. | Datetime format should be YYYY-MM-DD HH:MM:SS+00:00 (UTC) |
| med_name | VARCHAR | Name of the medication ordered | No restriction |
| med_category | VARCHAR | Maps med_name to a list of permissible medication names | Combined CDE of medication_admin_continuous and medication_admin_intermittent |
| med_group | VARCHAR | Limited number of medication groups identified by the CLIF consortium | No restriction |
| med_order_status_name | VARCHAR | Status of the medication order, e.g. held, given | No restriction |
| med_order_status_category | VARCHAR | Maps med_order_status_name to a standardized list of medication order statuses | Under-development |
| med_route_name | VARCHAR | Route of administration for the medication | No restriction. Examples include Oral, Intravenous |
| med_dose | DOUBLE | Dosage of the medication ordered | Numeric |
| med_dose_unit | VARCHAR | Unit of measurement for the medication dosage | Examples include mg, mL, units |
| med_frequency | VARCHAR | Frequency with which the medication is administered, as per the order | Examples include Once Daily, Every 6 hours |
| prn | BOOLEAN | Indicates whether the medication is to be given as needed (PRN) | 0 = No, 1 = Yes |
Example:
| hospitalization_id | med_order_id | order_start_dttm | order_end_dttm | ordered_dttm | med_name | med_category | med_group | med_order_status_name | med_order_status_category | med_route_name | med_dose | med_dose_unit | med_frequency | prn |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 12345 | 456789 | 2023-10-01 14:00:00+00:00 UTC | 2023-10-02 14:00:00+00:00 UTC | 2023-10-01 13:30:00+00:00 UTC | Norepinephrine 8 mg/250 mL | norepinephrine | vasoactives | active | ongoing | Intravenous | 8.0 | mg/mL | Continuous | 0 |
| 12346 | 456790 | 2023-10-01 16:00:00+00:00 UTC | 2023-10-02 10:00:00+00:00 UTC | 2023-10-01 15:30:00+00:00 UTC | Vancomycin 1 g IV | vancomycin | antibiotics | active | ongoing | Intravenous | 1.0 | g | Every 12 hours | 0 |
| 12347 | 456791 | 2023-10-02 08:00:00+00:00 UTC | 2023-10-03 08:00:00+00:00 UTC | 2023-10-02 07:30:00+00:00 UTC | Furosemide 40 mg IV | furosemide | diuretics | discontinued | discontinued | Intravenous | 40.0 | mg | Once Daily | 0 |
| 12348 | 456792 | 2023-10-02 12:00:00+00:00 UTC | 2023-10-02 18:00:00+00:00 UTC | 2023-10-02 11:45:00+00:00 UTC | Insulin Regular 100 units/mL SC | insulin | endocrine | held | held | Subcutaneous | 100.0 | units/mL | As Needed | 1 |
| 12349 | 456793 | 2023-10-03 08:00:00+00:00 UTC | 2023-10-03 20:00:00+00:00 UTC | 2023-10-03 07:30:00+00:00 UTC | Acetaminophen 1 g PO | acetaminophen | analgesics | active | ongoing | Oral | 1.0 | g | Every 6 hours | 0 |
| 12350 | 456794 | 2023-10-03 10:00:00+00:00 UTC | 2023-10-03 18:00:00+00:00 UTC | 2023-10-03 09:45:00+00:00 UTC | Heparin 5,000 units SC | heparin | anticoagulant | active | ongoing | Subcutaneous | 5000.0 | units | Every 8 hours | 0 |
| 12351 | 456795 | 2023-10-03 14:00:00+00:00 UTC | 2023-10-03 22:00:00+00:00 UTC | 2023-10-03 13:30:00+00:00 UTC | Morphine Sulfate 2 mg IV | morphine | analgesics | active | ongoing | Intravenous | 2.0 | mg | As Needed | 1 |
| 12352 | 456796 | 2023-10-03 20:00:00+00:00 UTC | 2023-10-04 08:00:00+00:00 UTC | 2023-10-03 19:45:00+00:00 UTC | Dexamethasone 10 mg IV | dexamethasone | steroids | active | ongoing | Intravenous | 10.0 | mg | Once Daily | 0 |
microbiology nonculture Concept
The microbiology non-culture table is a wide longitudinal table that captures the order and result times of non-culture microbiology tests, the type of fluid collected, the component of the test, and the result of the test.
| Column | Type | Description | Permissible Values |
|---|---|---|---|
| patient_id | VARCHAR | Unique identifier for each patient, presumed to be a distinct individual. | No restriction |
| hospitalization_id | VARCHAR | ID variable for each patient encounter | No restriction |
| result_dttm | DATETIME | Date and time when the non-culture result was obtained. All datetime variables must be timezone-aware and set to UTC. | Datetime format should be YYYY-MM-DD HH:MM:SS+00:00 |
| collect_dttm | DATETIME | Date and time when the sample was collected. All datetime variables must be timezone-aware and set to UTC. | Datetime format should be YYYY-MM-DD HH:MM:SS+00:00 |
| order_dttm | DATETIME | Date and time when the test was ordered. All datetime variables must be timezone-aware and set to UTC. | Datetime format should be YYYY-MM-DD HH:MM:SS+00:00 |
| fluid_name | VARCHAR | Name of the fluid sample. | No restriction |
| fluid_category | VARCHAR | Fluid categories defined according to the NIH common data elements. | CDE NIH Infection Site |
| method_name | VARCHAR | Original method names from the source data. | No restriction |
| method_category | VARCHAR | Maps method_name to a standardized list of method categories. | pcr |
| micro_order_name | VARCHAR | String name of microbiology non-culture test. | No restriction |
| organism_category | VARCHAR | Maps the organism name in micro_order_name to the standardized list of organisms under the structure of genus species. | Organism species. Check this file for examples. |
| organism_group | VARCHAR | Maps organism_category to the standardized list of organisms under the NIH CDE structure. | CDE NIH Organism |
| result_name | VARCHAR | Result name from the raw data. | No restriction |
| result_category | VARCHAR | Category of the test result. | Check list of result categories |
| reference_low | DOUBLE | Reference low value. | No restriction |
| reference_high | DOUBLE | Reference high value. | No restriction |
| result_units | VARCHAR | Unit of the test result. | No restriction |
| lab_loinc_code | VARCHAR | LOINC code. | No restriction |
Example:
| patient_id | hospitalization_id | order_dttm | collect_dttm | result_dttm | fluid_name | fluid_category | method_name | method_category | micro_order_name | organism_category | organism_group | result_name | result_category | reference_low | reference_high | result_units | lab_loinc_code |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 12121 | 2025-06-15 09:05:00+00:00 | 2025-06-15 09:30:00+00:00 | 2025-06-15 13:45:00+00:00 | BLOOD | blood/buffy coat | PCR | pcr | neisseria quantitative pcr, blood | neisseria_sp | neisseria (gonorrhoea, meningitidis, other species) | 100,000 copies/uL of neisseria detected | detected | copies/mL | 39528-5 | ||
| 2 | 32332 | 2025-06-16 11:15:00+00:00 | 2025-06-16 11:40:00+00:00 | 2025-06-16 15:25:00+00:00 | cerebrospinal fluid | meninges and csf | PCR | pcr | csf hsv pcr | herpes_simplex_virus | herpes simplex (hsv1, hsv2) | no herspes simplex DNA measured | not_detected | IU/mL | 16954-2 | ||
| 2 | 32332 | 2025-06-17 10:00:00+00:00 | 2025-06-17 10:20:00+00:00 | 2025-06-17 14:05:00+00:00 | feces | feces/stool | PCR | pcr | stool c. diff toxin | clostridioides_difficile | clostridium difficile | default in test for C. difficile toxin analysis | indeterminate | copies/mL | 34712-0 |
patient diagnosis Concept
The patient_diagnosis table provides a record of all diagnoses assigned to a patient.
| Column | Type | Description | Permissible Values |
|---|---|---|---|
| patient_id | VARCHAR | Unique identifier for each patient, presumed to be a distinct individual. | No restriction |
| hospitalization_id | VARCHAR | ID variable for each patient encounter | No restriction |
| diagnosis_code | VARCHAR | ICD-10-CM from clinical documentation | Valid ICD-10-CM code |
| diagnosis_code_format | VARCHAR | ICD10CM (clinical data typically ICD-10 only) | ICD10CM |
| source_type | VARCHAR | Source of diagnosis: problem_list, medical_history, encounter_dx (optional) | problem_list, medical_history, encounter_dx |
| start_dttm | DATETIME | When condition started (user-defined) | Datetime format should be YYYY-MM-DD HH:MM:SS+00:00 (UTC) |
| end_dttm | DATETIME | When condition ended (NULL if ongoing) | Datetime format should be YYYY-MM-DD HH:MM:SS+00:00 (UTC) |
Example:
| patient_id | hospitalization_id | diagnosis_code | diagnosis_code_format | source_type | start_dttm | end_dttm |
|---|---|---|---|---|---|---|
| PAT1001 | HOSP1001 | I10 | ICD10CM | problem_list | 2024-01-01 08:00:00+00:00 | NULL |
| PAT1001 | HOSP1001 | E11.9 | ICD10CM | encounter_dx | 2024-01-01 08:00:00+00:00 | 2024-01-10 12:00:00+00:00 |
| PAT1002 | HOSP1002 | J18.9 | ICD10CM | medical_history | 2024-01-05 09:30:00+00:00 | NULL |
| PAT1003 | HOSP1003 | N17.9 | ICD10CM | encounter_dx | 2024-01-10 07:00:00+00:00 | 2024-01-15 10:00:00+00:00 |
place based index Concept
The place_based_index table is designed to store geospatial or community-level indices linked to a patient’s hospitalization.
| Column | Type | Description | Permissible Values |
|---|---|---|---|
| hospitalization_id | VARCHAR | ID variable for each patient encounter | No restriction |
| index_name | VARCHAR | The name of the index (e.g., Area Deprivation Index, Social Vulnerability Index). | No restriction |
| index_value | DOUBLE | The numerical value of the index for the given hospitalization. | Numeric |
| index_version | VARCHAR | Version of the index used (e.g., ADI 2019, SVI 2020). | No restriction |
Notes:
Source indices from validated and publicly available datasets.
Example:
| hospitalization_id | index_name | index_value | index_version |
|---|---|---|---|
| 1001 | Area Deprivation Index | 85.2 | ADI 2019 |
| 1001 | Social Vulnerability Index | 0.72 | SVI 2020 |
| 1002 | Area Deprivation Index | 67.5 | ADI 2019 |
provider Concept
Continuous start stop record of every provider who cared for the patient.
| Column | Type | Description | Permissible Values |
|---|---|---|---|
| hospitalization_id | VARCHAR | ID variable for each patient encounter | No restriction |
| provider_id | VARCHAR | Unique identifier for each provider. This represents individual healthcare providers | No restriction |
| start_dttm | DATETIME | Date and time when the provider’s care or involvement in the patient’s case began. All datetime variables must be timezone-aware and set to UTC. | Datetime format should be YYYY-MM-DD HH:MM:SS+00:00 (UTC) |
| stop_dttm | DATETIME | Date and time when the provider’s care or involvement in the patient’s case ended. All datetime variables must be timezone-aware and set to UTC. | Datetime format should be YYYY-MM-DD HH:MM:SS+00:00 (UTC) |
| provider_role_name | VARCHAR | The original string describing the role or specialty of the provider during the hospitalization | No restriction |
| provider_role_category | VARCHAR | Maps provider_role_name to list of standardized provider roles | Under development |
Example:
| hospitalization_id | start_dttm | stop_dttm | provider_role_name | provider_role_category |
|---|---|---|---|---|
| 1001014 | 2023-05-01 08:00:00+00:00 UTC | 2023-05-01 20:00:00+00:00 UTC | Attending Physician | Attending |
| 1001014 | 2023-05-01 08:00:00+00:00 UTC | 2023-05-02 08:00:00+00:00 UTC | Resident Physician | Resident |
| 1001014 | 2023-05-01 08:00:00+00:00 UTC | 2023-05-03 08:00:00+00:00 UTC | Nurse Practitioner | Nurse Practitioner |
| 1002025 | 2023-06-10 09:00:00+00:00 UTC | 2023-06-10 21:00:00+00:00 UTC | Critical Care Specialist | Critical Care |
| 1002025 | 2023-06-10 09:00:00+00:00 UTC | 2023-06-11 09:00:00+00:00 UTC | Respiratory Therapist | Respiratory Therapy |
| 1003036 | 2023-07-15 07:30:00+00:00 UTC | 2023-07-15 19:30:00+00:00 UTC | Attending Physician | Attending |
| 1003036 | 2023-07-15 07:30:00+00:00 UTC | 2023-07-16 07:30:00+00:00 UTC | Charge Nurse | Nurse |
| 1004047 | 2023-08-20 10:00:00+00:00 UTC | 2023-08-20 22:00:00+00:00 UTC | Physical Therapist | Therapy |
therapy details Concept
The therapy_details table is a wide longitudinal table that captures the details of therapy sessions. The table is designed to capture and categorize the most common therapy elements used in the ICU.
| Column | Type | Description | Permissible Values |
|---|---|---|---|
| hospitalization_id | VARCHAR | ID variable for each patient encounter | No restriction |
| session_start_dttm | DATETIME | Date and time when the therapy session started. All datetime variables must be timezone-aware and set to UTC. | Datetime format should be YYYY-MM-DD HH:MM:SS+00:00 (UTC) |
| therapy_element_name | VARCHAR | Name of the therapy element. | No restriction |
| therapy_element_category | VARCHAR | Category of the therapy element. | No restriction |
| therapy_element_value | VARCHAR | Value associated with the therapy element. | No restriction |
Example:
| hospitalization_id | session_start_dttm | therapy_element_name | therapy_element_category | therapy_element_value |
|---|---|---|---|---|
| 1001 | 2024-01-01 08:00:00+00:00 UTC | Physical Therapy | Rehabilitation | 45.0 |
| 1001 | 2024-01-01 10:00:00+00:00 UTC | Respiratory Therapy | Respiratory Support | 3.0 |
| 1002 | 2024-01-05 09:30:00+00:00 UTC | Occupational Therapy | Rehabilitation | 60.0 |
| 1002 | 2024-01-05 11:00:00+00:00 UTC | Speech Therapy | Rehabilitation | 30.0 |
| 1003 | 2024-01-10 07:00:00+00:00 UTC | Ventilation Support | Respiratory Support | 2.5 |
transfusion Concept
This table provides detailed information about transfusion events linked to specific hospitalizations.
| Column | Type | Description | Permissible Values |
|---|---|---|---|
| hospitalization_id | VARCHAR | ID variable for each patient encounter | No restriction |
| transfusion_start_dttm | DATETIME | The date and time the transfusion of the blood component began. All datetime variables must be timezone-aware and set to UTC. | Datetime format should be YYYY-MM-DD HH:MM:SS+00:00 (UTC) |
| transfusion_end_dttm | DATETIME | The date and time the transfusion of the blood component ended. All datetime variables must be timezone-aware and set to UTC. | Datetime format should be YYYY-MM-DD HH:MM:SS+00:00 (UTC) |
| component_name | VARCHAR | The name of the blood component transfused. | E.g., Red Blood Cells, Plasma, Platelets |
| attribute_name | VARCHAR | Attributes describing modifications to the component. | E.g., Leukocyte Reduced, Irradiated |
| volume_transfused | DOUBLE | The volume of the blood component transfused. | Numeric, e.g., 300 |
| volume_units | VARCHAR | The unit of measurement for the transfused volume. | E.g., mL |
| product_code | VARCHAR | ISBT 128 Product Description Code representing the specific blood product. | E.g., E0382 |
Example:
| hospitalization_id | transfusion_start_dttm | transfusion_end_dttm | component_name | attribute_name | volume_transfused | volume_units | product_code |
|---|---|---|---|---|---|---|---|
| 123456 | 2024-12-03 08:30:00+00:00 UTC | 2024-12-03 10:00:00+00:00 UTC | Red Blood Cells | Leukocyte Reduced | 300 | mL | E0382 |
| 789012 | 2024-12-04 14:00:00+00:00 UTC | 2024-12-04 16:30:00+00:00 UTC | Platelets | Irradiated | 250 | mL | P0205 |
| 456789 | 2024-12-05 12:15:00+00:00 UTC | 2024-12-05 13:45:00+00:00 UTC | Plasma | 200 | mL | F0781 |
validated diagnosis Concept
The validated_diagnosis table captures clinician-validated diagnostic labels for research purposes. This table is designed for studies requiring confirmed diagnoses through chart review or consensus adjudication, enabling high-quality phenotyping for research cohorts.
| Column | Type | Description | Permissible Values |
|---|---|---|---|
| hospitalization_id | VARCHAR | ID variable for each patient encounter. | No restriction |
| diagnosis_name | VARCHAR | Free-text name of the diagnosis (clinician-entered or source name). | No restriction |
| diagnosis_category | VARCHAR | Maps diagnosis_name to a standardized list of diagnosis categories. | e.g., sepsis, arf, aki, stroke, etc. |
| diagnosis_start_dttm | DATETIME | Date and time when the diagnosis is deemed clinically active. | Datetime format should be YYYY-MM-DD HH:MM:SS+00:00 (UTC) |
| diagnosis_end_dttm | DATETIME | Date and time when the diagnosis was resolved or deemed no longer active (optional). | Datetime format should be YYYY-MM-DD HH:MM:SS+00:00 (UTC) |
| validation_method | VARCHAR | Method of clinician validation used for this label. | manual_chart_review, consensus_panel, automated_with_review |
| diagnosis_status | VARCHAR | Final reviewer determination. | confirmed, ruled_out, uncertain |
| reviewer_id | VARCHAR | Anonymized ID of the clinician or reviewer responsible for this label. | No restriction |
| review_timestamp | DATETIME | Time when the label was finalized or adjudicated. | Datetime format should be YYYY-MM-DD HH:MM:SS+00:00 (UTC) |
Notes:
- This table is intended for research diagnoses that have undergone validation, not routine clinical diagnoses
- The
diagnosis_categoryshould map to a controlled vocabulary appropriate for the research context - Multiple reviewers can validate the same diagnosis by creating separate rows with different
reviewer_idvalues
Example:
| hospitalization_id | diagnosis_name | diagnosis_category | diagnosis_start_dttm | diagnosis_end_dttm | validation_method | diagnosis_status | reviewer_id | review_timestamp |
|---|---|---|---|---|---|---|---|---|
| 12345 | Septic shock | sepsis | 2024-12-01 08:00:00+00:00 UTC | 2024-12-03 14:00:00+00:00 UTC | manual_chart_review | confirmed | REV001 | 2024-12-15 10:30:00+00:00 UTC |
| 12345 | Acute kidney injury | aki | 2024-12-01 12:00:00+00:00 UTC | NULL | consensus_panel | confirmed | REV002 | 2024-12-16 09:00:00+00:00 UTC |
| 67890 | Acute respiratory failure | arf | 2024-11-28 06:00:00+00:00 UTC | 2024-11-30 18:00:00+00:00 UTC | automated_with_review | uncertain | REV001 | 2024-12-10 14:15:00+00:00 UTC |
| 54321 | Ischemic stroke | stroke | 2024-12-05 02:30:00+00:00 UTC | NULL | manual_chart_review | ruled_out | REV003 | 2024-12-18 11:45:00+00:00 UTC |
Future Proposed Tables
These are tables without any defined structure that the consortium has not yet committed to implementing.
Clinical Decision Support: This table will capture the actions of clinical decision support tools embedded in the EHR. The table will have the following fields: cds_name, cds_category, cds_value, cds_trigger_ddtm.